Nomenclature
Short Name:
BUBR1
Full Name:
Mitotic checkpoint serine-threonine-protein kinase BUB1 beta
Alias:
- BUB1 budding uninhibited by benzimidazoles 1 homologue beta (yeast)
- EC 2.7.11.1
- HBUBR1
- Kinase BubR1
- MAD3/BUB1-related protein kinase
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
BUB
SubFamily:
NA
Structure
Mol. Mass (Da):
119545
# Amino Acids:
1050
# mRNA Isoforms:
3
mRNA Isoforms:
121,386 Da (1064 AA; O60566-3); 119,545 Da (1050 AA; O60566); 105,890 Da (933 AA; O60566-2)
4D Structure:
Interacts with CENPE, CENPF, mitosin, PLK1 and BUB3. Part of a complex containing BUB3, CDC20 and BUB1B. Interacts with anaphase-promoting complex/cyclosome (APC/C). Interacts with CASC5.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 423 | 465 | Coiled-coil |
| 766 | 1033 | Pkinase |
| 62 | 213 | CD1 |
| 55 | 179 | MAD3 |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K250 (N6), K451.
Serine phosphorylated:
S39, S83, S367, S384, S435, S521, S537, S543, S574, S633, S665, S670, S676, S683, S697, S720, S733, S797, S884, S1043.
Sumoylated:
K250.
Threonine phosphorylated:
T40, T54, T315, T368, T434, T471, T600, T608, T620+, T676, T680, T710, T792+, T1008+, T1042.
Tyrosine phosphorylated:
Y404, Y660, Y766.
Ubiquitinated:
K113, K210, K250, K475, K539, K629, K995, K1040.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1880
16
3856
 0.9
0.9
17
10
18
 6
6
115
20
221
 16
16
306
72
817
 23
23
439
14
449
 10
10
192
44
296
 15
15
286
19
466
 87
87
1628
44
4755
 19
19
364
10
362
 2
2
38
44
39
 3
3
47
36
89
 22
22
411
117
567
 5
5
87
34
111
 0.8
0.8
15
9
11
 3
3
54
33
102
 0.7
0.7
14
8
11
 3
3
65
123
363
 7
7
126
29
339
 1.4
1.4
26
65
54
 23
23
428
56
473
 3
3
52
32
97
 3
3
60
34
83
 5
5
86
24
104
 17
17
324
30
250
 19
19
359
32
351
 63
63
1184
54
3474
 5
5
92
37
75
 4
4
76
30
139
 3
3
51
30
80
 4
4
68
14
46
 26
26
484
18
350
 34
34
642
21
707
 77
77
1455
55
1839
 43
43
807
26
699
 13
13
242
22
229
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 89.2
89.2
90.4
100 -
-
-
97 -
-
-
89 -
-
-
95 83.2
83.2
87.1
89 -
-
-
- 75.8
75.8
84.5
77 -
-
-
78 -
-
-
- -
-
-
- -
-
-
56 -
-
-
59 -
-
-
47 -
-
-
- 21.3
21.3
37.2
- -
-
-
- -
-
-
- 23.6
23.6
39.6
- -
-
-
- -
-
-
- -
-
-
35 -
-
-
34 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC20 - Q12834 |
| 2 | BUB3 - O43684 |
| 3 | CENPE - Q02224 |
| 4 | CDC16 - Q13042 |
| 5 | CDC27 - P30260 |
| 6 | SNCG - O76070 |
| 7 | BRCA2 - P51587 |
| 8 | ANAPC7 - Q9UJX3 |
| 9 | APC - P25054 |
| 10 | ZWINT - O95229 |
| 11 | SMAD2 - Q15796 |
| 12 | HDAC1 - Q13547 |
| 13 | AP4B1 - Q9Y6B7 |
| 14 | AP2B1 - P63010 |
| 15 | AP3B1 - O00203 |
Regulation
Activation:
Activated by CENPE, which increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
Induced during mitosis.
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| TTK | P33981 | S435 | REAELLTSAEKRAEM | |
| TTK | P33981 | S543 | LSEKKNKSPPADPPR | |
| CDK1 | P06493 | S543 | LSEKKNKSPPADPPR | |
| TTK | P33981 | S670 | TLSIKKLSPIIEDSR | ? |
| CDK1 | P06493 | S670 | TLSIKKLSPIIEDSR | ? |
| PLK1 | P53350 | T676 | LSPIIEDSREATHSS | ? |
| PLK1 | P53350 | T680 | IEDSREATHSSGFSG | ? |
| PLK1 | P53350 | T792 | PRNSAELTVIKVSSQ | + |
| PLK1 | P53350 | T1008 | LNANDEATVSVLGEL | + |
| TTK | P33981 | S1043 | WKVGKLTSPGALLFQ | |
| CDK1 | P06493 | S1043 | WKVGKLTSPGALLFQ |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
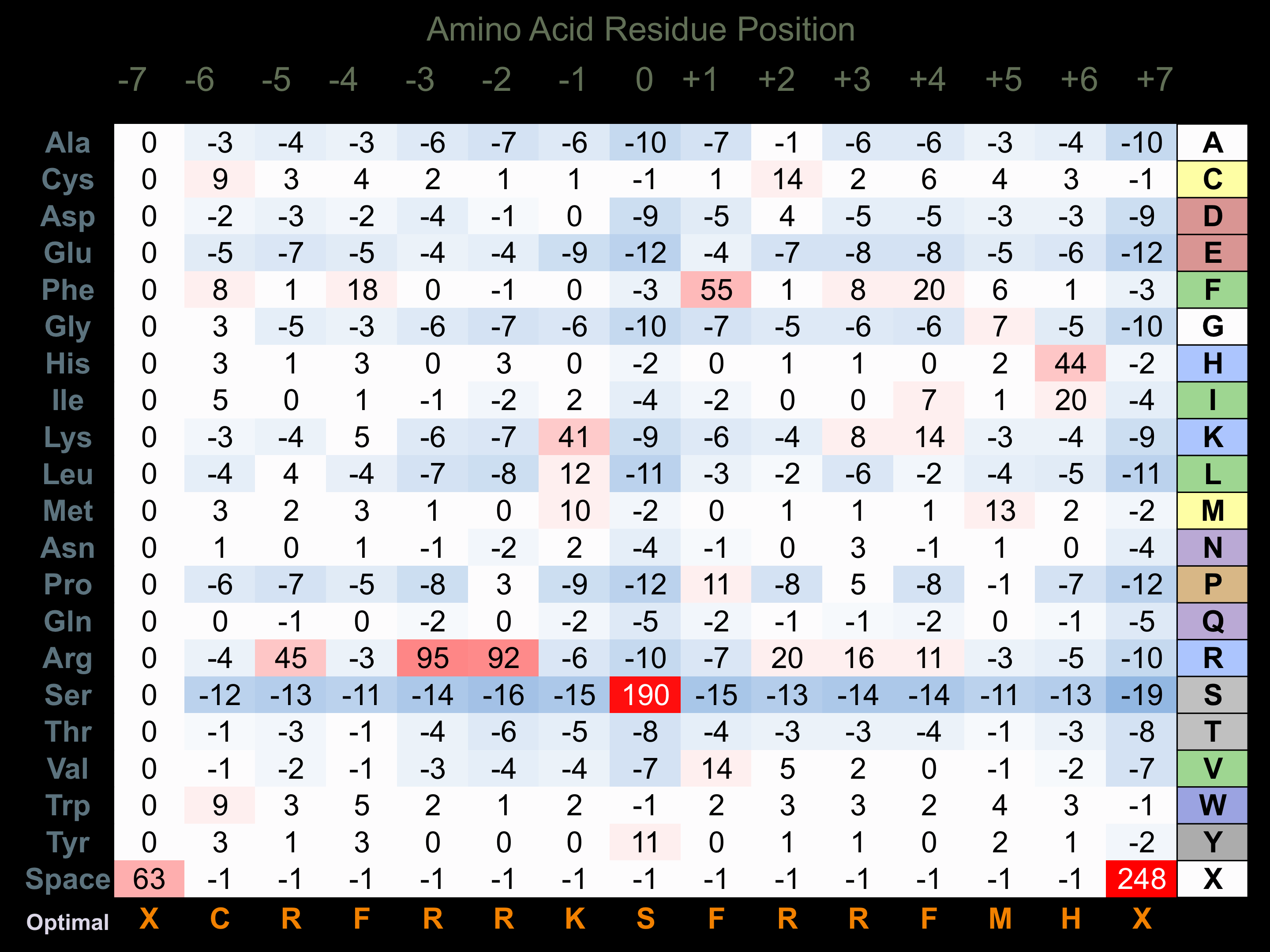
Matrix Type:
Derived from alignment of 33 peptides phosphorylated by recombinant BUBR1 in vitro tested in-house by Kinexus.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer, neurological, and muscle disorders
Specific Diseases (Non-cancerous):
Mosaic variegated aneuploidy syndrome 1; Premature chromatid separation trait
Comments:
In five families with Mosaic variegated aneuploidy syndrome 1 (MVA1), including two with embryonal rhabdomyosarcoma, truncating and missense mutations of BUBR1 have been observed, including an R550Q mutation. MVA1 is a rare, recessive condition characterized by growth retardation, microcephaly, childhood cancer and constitutional mosaic aneuploides with chromosomal gains and losses. It is lkely that loss of function of BUBR1 contributions to MVA1.
Specific Cancer Types:
Colorectal cancer (CRC); Plexiform neurofibromas (PNF); Colorectal cancer (CRC), somatic
Comments:
BUBR1 may be a tumour suppressor protein (TSP). An R36Q mutation is linked with premature chromatid separation trait (PCS), which is an autosomal dominant disease. An R550Q mutation is found in Mosaic variegated aneuploidy syndrome 1 (MVA1), which is a severe developmental disease with characterized mosaic aneuploidies. BUBR1 acts as a tumour suppresser, and loss of function mutations are associated with tumour formation.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +71, p<0.01); Bladder carcinomas (%CFC= +187, p<0.006); Brain glioblastomas (%CFC= +150, p<0.016); Brain oligodendrogliomas (%CFC= +208, p<0.011); Breast epithelial carcinomas (%CFC= +193, p<0.068); Breast non-basal-like cancer (BLC) (%CFC= +80, p<0.0001); Breast sporadic basal-like cancer (BLC) (%CFC= +94, p<0.0001); Cervical cancer (%CFC= -53, p<0.0001); Cervical cancer stage 1B (%CFC= -64, p<0.047); Cervical cancer stage 2A (%CFC= -51, p<0.09); Classical Hodgkin lymphomas (%CFC= +164, p<0.002); Colon mucosal cell adenomas (%CFC= +158, p<0.0001); Head and neck squamous cell carcinomas (HNSCC) (%CFC= +59, p<0.0002); Large B-cell lymphomas (%CFC= +365, p<0.006); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +79, p<0.019); Oral squamous cell carcinomas (OSCC) (%CFC= +102, p<0.032); Ovary adenocarcinomas (%CFC= +747, p<0.0002); Skin squamous cell carcinomas (%CFC= +54, p<0.029); and Vulvar intraepithelial neoplasia (%CFC= +218, p<0.0002). The COSMIC website notes an up-regulated expression score for BUBR1 in diverse human cancers of 472, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 16 for this protein kinase in human cancers was 0.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. A K795R mutation can inhibit the phosphotransferase activity of BUBR1, but does not abolish its ability to inhibit the anaphase promoting complex (APC)/cycolsome.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 25634 diverse cancer specimens. This rate is only -34 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.22 % in 805 skin cancers tested; 0.18 % in 589 stomach cancers tested; 0.18 % in 1152 large intestine cancers tested; 0.09 % in 1270 liver cancers tested; 0.08 % in 1942 lung cancers tested.
Frequency of Mutated Sites:
None > 6 in 20,872 cancer specimens
Comments:
Only 4 deletions, no insertions or complex mutations are noted on the COSMIC website.

