Nomenclature
Short Name:
BCR
Full Name:
Breakpoint cluster region protein
Alias:
- ALL
- BCR
- D22S662
- PHL
- Renal carcinoma antigen NY-REN-26
- BCR1
- Breakpoint cluster region
- CML
- D22S11
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
BCR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
142819
# Amino Acids:
1271
# mRNA Isoforms:
2
mRNA Isoforms:
142,819 Da (1271 AA; P11274); 137,729 Da (1227 AA; P11274-2)
4D Structure:
Homotetramer. Interacts with PDZK1. May interact with CCPG1
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
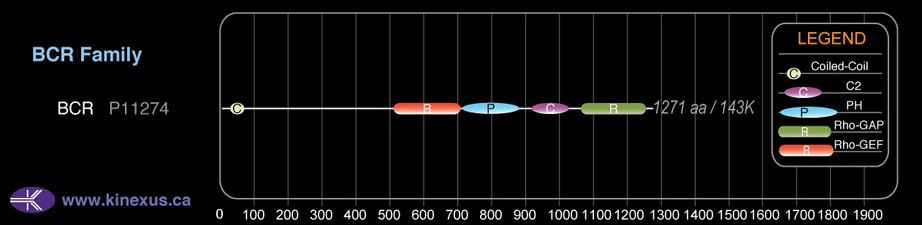
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K194, K213.
Serine phosphorylated:
S18, S88, S122, S139, S201, S202, S205, S215, S221, S222, S227, S235, S236, S239, S267, S281, S299, S301, S303, S317, S320, S340, S351, S354, S356, S374, S377, S429, S459, S461, S462, S463, S467, S468, S473, S488, S515, S540, S572, S626, S638, S653, S698, S843, S876, S885, S894, S906, S919, S1030, S1035, S1039, S1264.
Threonine phosphorylated:
T130, T302, T310, T329, T358, T359, T410, T432, T634, T635, T641, T651, T693, T845, T898, T921, T1018, T1049.
Tyrosine phosphorylated:
Y58, Y177+, Y231, Y246+, Y276, Y279, Y283, Y328+, Y360-, Y436, Y455, Y513, Y554, Y561, Y591, Y598, Y644, Y751, Y756, Y844, Y852, Y910, Y1069, Y1089.
Ubiquitinated:
K486, K494, K583.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 31
31
1008
29
1163
 3
3
84
22
71
 5
5
152
45
102
 11
11
357
147
424
 21
21
690
28
660
 3
3
97
80
100
 18
18
577
50
665
 17
17
536
92
1426
 10
10
337
20
365
 7
7
224
172
181
 4
4
129
78
122
 14
14
457
290
573
 5
5
165
67
132
 2
2
64
18
46
 5
5
164
73
153
 3
3
105
19
80
 3
3
97
491
95
 6
6
177
62
151
 2
2
57
147
47
 17
17
557
112
570
 8
8
257
71
326
 4
4
130
79
118
 6
6
186
63
108
 4
4
124
63
93
 6
6
181
72
163
 18
18
568
102
1131
 5
5
148
73
133
 6
6
186
61
141
 4
4
128
61
98
 2
2
56
28
23
 21
21
667
36
445
 100
100
3215
42
7182
 5
5
173
107
756
 18
18
593
62
571
 2
2
77
53
95
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
100 -
-
-
99 -
-
-
94 -
-
-
- 91.8
91.8
93.7
94 -
-
-
- 93.9
93.9
95.8
94 23.1
23.1
36
93 -
-
-
- 41.9
41.9
50.3
- -
-
-
87 45.4
45.4
56.3
77 -
-
-
79 -
-
-
- -
-
-
36 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | GRB2 - P62993 |
| 2 | ABL1 - P00519 |
| 3 | CRKL - P46109 |
| 4 | HCK - P08631 |
| 5 | PIK3CG - P48736 |
| 6 | FES - P07332 |
| 7 | PTPN11 - Q06124 |
| 8 | MLLT4 - P55196 |
| 9 | PTPN6 - P29350 |
| 10 | SOS1 - Q07889 |
| 11 | CFTR - P13569 |
| 12 | SHC1 - P29353 |
| 13 | HOXA9 - P31269 |
| 14 | IRS1 - P35568 |
| 15 | YWHAZ - P63104 |
Regulation
Activation:
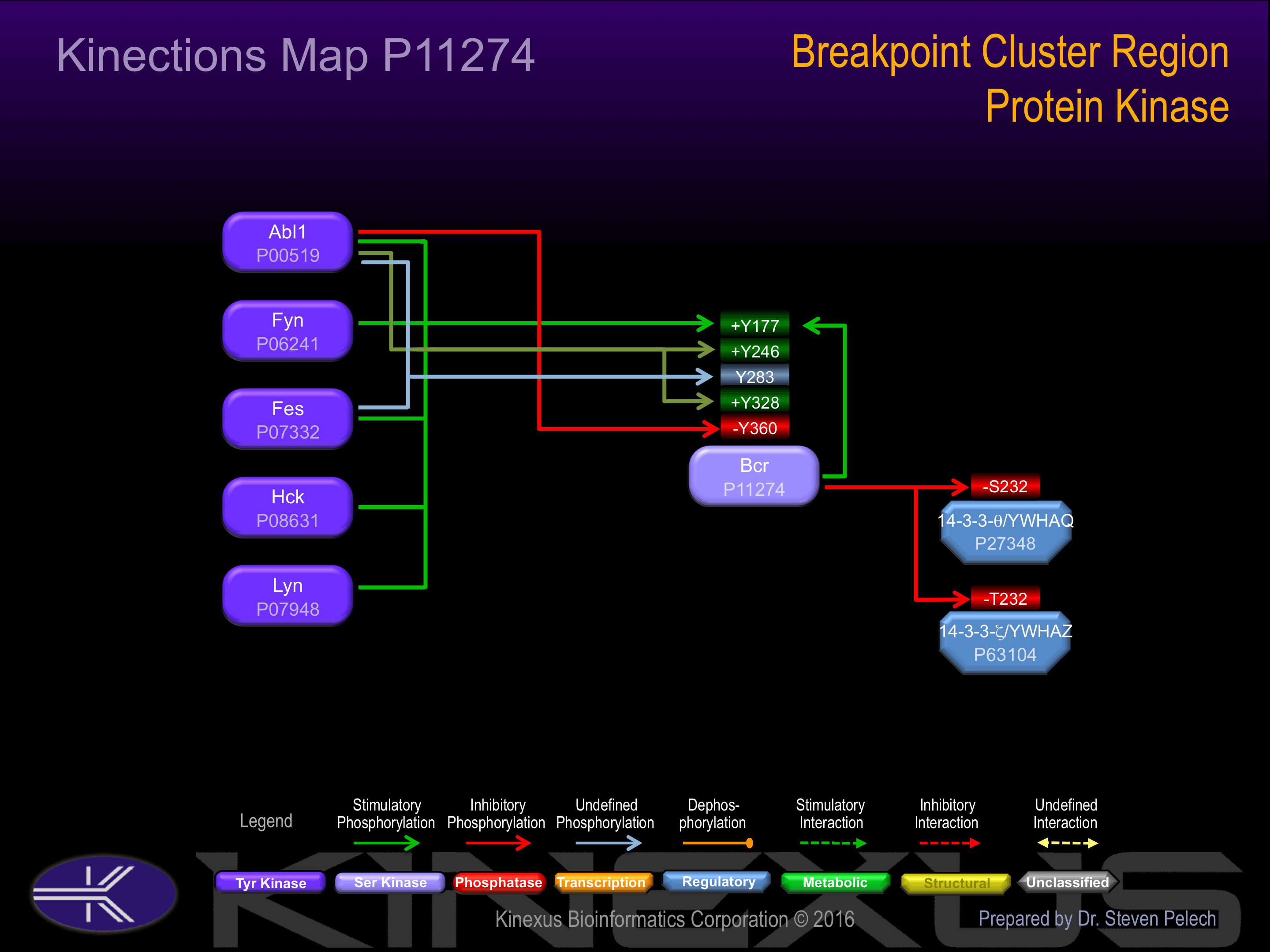
BCR promotes the exchange of Rac or Cdc42-bound GDP by GTP, thereby activating them.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| LYN | P07948 | Y177 | ADAEKPFYVNVEFHH | + |
| FES | P07332 | Y177 | ADAEKPFYVNVEFHH | + |
| FYN | P06241 | Y177 | ADAEKPFYVNVEFHH | + |
| ABL | P00519 | Y177 | ADAEKPFYVNVEFHH | + |
| BCR | P11274 | Y177 | ADAEKPFYVNVEFHH | + |
| HCK | P08631 | Y177 | ADAEKPFYVNVEFHH | + |
| FES | P07332 | Y246 | SCGVDGDYEDAELNP | + |
| ABL | P00519 | Y246 | SCGVDGDYEDAELNP | + |
| FES | P07332 | Y283 | YQPYQSIYVGGMMEG | |
| ABL | P00519 | Y283 | YQPYQSIYVGGMMEG | |
| ABL | P00519 | Y328 | FEDCGGGYTPDCSSN | + |
| ABL | P00519 | Y360 | VSPSPTTYRMFRDKS | - |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, blood disorders
Specific Diseases (Non-cancerous):
Cryoglobulinemia; Xeroderma pigmentosum, group B; Distal 22q11.2 microdeletion syndrome
Comments:
Cryoglobulinemia is a disease characterized by the presence of cryoglobulins in the blood stream, which become solid at low temperatures leading to the blockage of blood flow through smaller vessels. Symptoms of this disease include difficulty breathing, tiredness, glomerulonephritis, joint and muscle pain, skin ulcers, and skin death. Xeroderma pigmentosum is a skin disease characterized by extreme sensitivity to ultraviolet (UV) light, which may be accompanied by neurological defects. This disease is inherited in an autosomal recessive manner. BCR (breakpoint cluster region) is a protein-serine/threonine kinase which also acts as a GTPase-activating protein for RAC1 and CDC42.
Specific Cancer Types:
Chronic myeloid leukemias (CML); Myeloproliferative disorder; Lymphoblastic leukemias; Precursor T-cell acute lymphoblastic leukemias (ALL); Acute lymphocytic leukemias (ALL); Philadelphia chromosome-negative chronic myeloid leukemias (CML); Pilomyxoid astrocytomas; Gastrointestinal stromal tumours; 8p11 myeloproliferative syndrome; Myeloproliferative disorder with eosinophilias; leukemias, acute lymphoblastic 3; leukemias; Myeloid leukemias
Comments:
BCR appears to be an oncoprotein (OP). The BCR gene is located on chromosome 22q11. 2, which is the site of a common breakpoint for chromosomal translocations commonly found in specific human cancers. These chromosomal translocations are observed in chronic myeloid leukemia (CML) and acute lymphocytic leukemia (ALL) and produce two alternative forms of the Philadelphia chromosome, in which different sets of exons from the BCR gene are joined to a common group of exons from the ABL1 gene. This creates two chimeric oncogene products referred to as p210(BCR-ABL) and p185(BCR-ABL), which both display oncogenic tyrosine kinase activity. The p210(BCR-ABL) is the more common form and is found in more than 90% of patients with CML and in 25-30% of adult and 2-10% of childhood cases of acute lymphoblastic leukemia. The twon chimeric protein products exhibit a gain-of-function in the ABL tyrosine kinase activity, which is predicted to be involved in the malignant process and development of the cancer. Approximately 10% of patients with ALL have the translocation t(9;22)(q34;q11), which is identical to the translocation observed in CML. The inhibtion of myeloid precursor cell differentiation is a key feature of the cancer progression in patients with CML that express the p210(BCR-ABL) oncoprotein. This differentiation arrest is mediated through the transcriptional suppression of the granulocyte-stimulating factor receptor (CSF3R) gene through phosphorylation by the ABL kinase domain of the fusion protein. In animal studies, transplantation of a retrovirus encoding the p210(BCR-ABL) oncoprotein into irradiated immuno-deficient mice induced hematologic malignancies with similar features as human leukemia, specifically CML.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +47, p<0.002); Colon mucosal cell adenomas (%CFC= +51, p<0.0001); and Pituitary adenomas (aldosterone-secreting) (%CFC= +49, p<0.073). The COSMIC website notes an up-regulated expression score for BCR in diverse human cancers of 423, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 96 for this protein kinase in human cancers was 1.6-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25580 diverse cancer specimens. This rate is only -22 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.29 % in 805 skin cancers tested; 0.24 % in 1093 large intestine cancers tested; 0.16 % in 589 stomach cancers tested; 0.14 % in 602 endometrium cancers tested; 0.12 % in 864 upper aerodigestive tract cancers tested; 0.06 % in 1753 lung cancers tested; 0.04 % in 2161 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,674 cancer specimens
Comments:
Only 4 deletions, 2 insertions, and no complex mutations are noted on the COSMIC website.