Nomenclature
Short Name:
CDK11B
Full Name:
Cell division protein kinase 11B
Alias:
- EC=2.7.11.22
- Cell division cycle 2-like protein kinase 1
- PK58
- Galactosyltransferase-associated protein kinase p58/GTA
- CLK-1
- p58 CLK-1
- CDK11
- PITSLREA
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
PITSLRE subfamily
Structure
Mol. Mass (Da):
92,707
# Amino Acids:
795
# mRNA Isoforms:
10
mRNA Isoforms:
92,707 Da (795 AA; P21127); 91,331 Da (782 AA; P21127-2); 91,218 Da (781 AA; P21127-3); 90,256 Da (772 AA; P21127-8); 87,138 Da (748 AA; P21127-9); 86,063 Da (738 AA; P21127-6); 63,937 Da (565 AA; P21127-10); 59,273 Da (526 AA; P21127-5); 51,925 Da (461 AA; P21127-4); 49,555 Da (439 AA; P21127-12)
4D Structure:
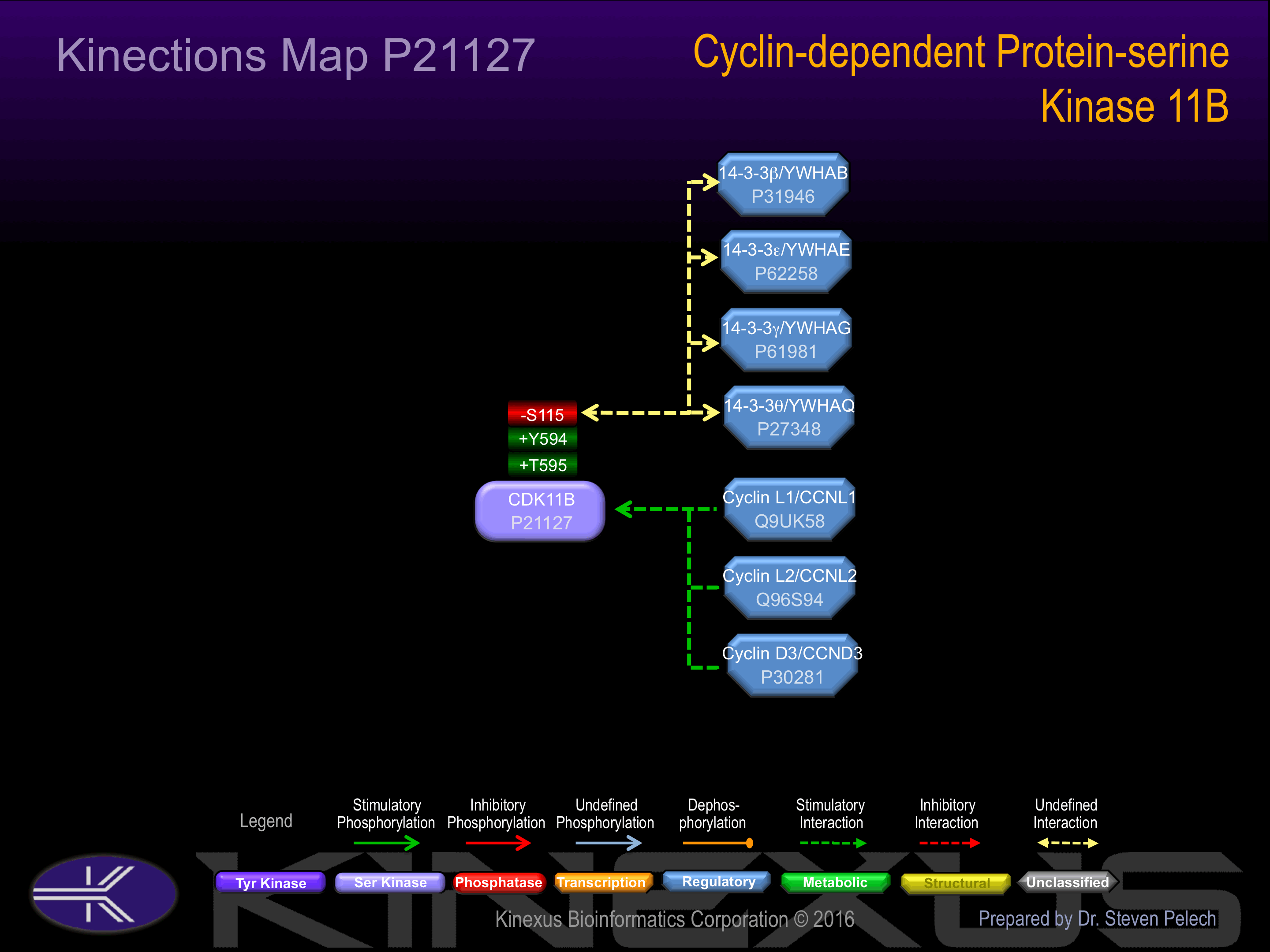
The cleaved p110 isoform, p110C, binds to the serine/threonine kinase PAK1 and RANBP9. p110C interacts with RNPS1. The p58 isoform but not the p110 isoforms or p110C interacts with CCND3. The p110 isoforms are found in large molecular weight complexes containing CCNL1 and SFRS7.
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 21 | 335 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K101, K135, K456, K458, K459, K467.
Serine phosphorylated:
S47, S65, S72, S113, S115-, S212, S229, S232, S234, S277, S283, S285, S381, S384, S410, S414, S434, S589, S752, S763, S785, S792.
Threonine phosphorylated:
T61, T395, T448, T595+, T600, T675, T751, T773, T778.
Tyrosine phosphorylated:
Y67, Y222, Y406, Y449, Y453, Y587, Y594+, Y762.
Ubiquitinated:
K20, K467, K577, K660, K672, K719, K741.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 26
26
715
6
700
 -
-
-
-
-

 -
-
-
-
-
 36
36
1002
6
1672
 -
-
-
-
-
 9
9
239
2
83
 8
8
231
2
49
 7
7
189
2
80
 -
-
-
-
-
 -
-
-
-
-
 5
5
135
2
65
 -
-
-
-
-
 4
4
107
3
94
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 5
5
139
2
12
 8
8
229
2
11
 -
-
-
-
-
 -
-
-
-
-
 9
9
256
2
34
 98
98
2697
4
1921
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 53
53
1465
12
91
 100
100
2765
6
6772
 -
-
-
-
-
 -
-
-
-
-
 7
7
185
9
133
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
98 65.6
65.6
66
99 -
-
-
94 -
-
-
98 94.7
94.7
96.4
94 -
-
-
- 95.2
95.2
97.2
92 50.1
50.1
52.7
93 -
-
-
- -
-
-
- 89.8
89.8
92.9
93 84.7
84.7
91.5
89 77.3
77.3
86.2
88 -
-
-
- 41.2
41.2
54.7
- 46.4
46.4
60.2
- 38.2
38.2
55.3
- 47
47
61.4
- -
-
-
- -
-
-
- -
-
-
40 -
-
-
49 20.6
20.6
28.1
- -
-
-
47.5
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CCNL1 - Q9UK58 |
| 2 | PAK1 - Q13153 |
| 3 | RNPS1 - Q15287 |
| 4 | EIF3F - O00303 |
| 5 | MYST2 - O95251 |
| 6 | PAK1 - Q13153 |
| 7 | YWHAE - P62258 |
| 8 | YWHAG - P61981 |
| 9 | CASP1 - P29466 |
| 10 | CASP3 - P42574 |
| 11 | CARD17 - Q5XLA6 |
| 12 | CDK7 - P50613 |
| 13 | SF3A2 - Q15428 |
| 14 | POLR2A - P24928 |
Regulation
Activation:
Based on homology with related cyclin-dependent kinases, phosphorylation at Thr-595 may activate the phosphotransferase activity of CDK11B.
Inhibition:
Based on homology with related cyclin-dependent kinases, phosphorylation at Thr-448 or Tyr-449 may inactivate the kinase.
Synthesis:
The p58 isoform is specifically induced in G2/M phase of the cell cycle.
Degradation:
NA
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| AT7519 | Kd = 8.4 nM | 11338033 | 22037378 | |
| AC1NS7CD | Kd = 98 nM | 5329665 | 295136 | 22037378 |
| SNS032 | Kd = 98 nM | 3025986 | 296468 | 18183025 |
| JNJ-7706621 | Kd = 150 nM | 5330790 | 191003 | 18183025 |
| Tozasertib | Kd = 290 nM | 5494449 | 572878 | 18183025 |
| Foretinib | Kd = 640 nM | 42642645 | 1230609 | 22037378 |
| Crizotinib | Kd = 760 nM | 11626560 | 601719 | 22037378 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Barasertib | Kd = 1.2 µM | 16007391 | 215152 | 18183025 |
| Linifanib | Kd = 1.3 µM | 11485656 | 223360 | 18183025 |
| AST-487 | Kd = 1.4 µM | 11409972 | 574738 | 18183025 |
| Pazopanib | Kd = 2.1 µM | 10113978 | 477772 | 18183025 |
| R547 | Kd = 2.3 µM | 6918852 | 22037378 |
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Neuroblastomas (NB); Endodermal sinus tumours; Childhood endodermal sinus tumours
Comments:
CDK11B may be a tumour suppressor protein (TSP). The p58 isoform of CDK11B is thought to function as an inhibitor of normal cell cycle progression and is well conserved across species, indicating a tumour suppressor function for the protein. Corresponding with this prediction, several loss-of-function mutations in the CDK11B gene have been associated with cancer. For example, at least 1 allele of the CDK11B gene was deleted or translocated in 18 out of 20 neuroblastoma cell lines analyzed. However, the CDK11B gene is located outside of the region of allelic loss of heterozygosity (LOH) commonly associated with neuroblastoma (1p36. 3-p36. 2), therefore the specific role of the CDK11B protein in the development of this cancer is unclear. In animal studies, mice haploinsufficient for CDK11 protein expression display increased rates of tumour development compared to wild-type controls, indicating a role for CDK11 the prevention of cancer development, most likely through a maintenance of genomic stability.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.02 % in 24433 diverse cancer specimens. This rate is -67 % lower than the average rate of 0.075 % calculated for human protein kinases in general. Such a low frequency of mutation in human cancers is consistent with this protein kinase playing a role as a tumour requiring protein (TRP).
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: D641E (5).
Comments:
Seven E322 to E324del EEE (in frame deletions) noted on the COSMIC website, but no other deletions, insertions or complex mutations.