Nomenclature
Short Name:
FLT4
Full Name:
Vascular endothelial growth factor receptor 3
Alias:
- EC 2.7.1.112
- VGFR3
- VGR3
- EC 2.7.10.1
- Fms-related tyrosine kinase 4
- PCL
- Tyrosine-protein kinase receptor FLT4
- VEGFR3
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
VEGFR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
145,599
# Amino Acids:
1298
# mRNA Isoforms:
3
mRNA Isoforms:
152,757 Da (1363 AA; P35916); 145,599 Da (1298 AA; P35916-1); 93,174 Da (830 AA; P35916-3)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N33, N104, N166, N251, N299, N411, N515, N527, N594, N683, N690, N758.
Serine phosphorylated:
S863, S871, S976, S986, S1163, S1235.
Threonine phosphorylated:
T517, T886.
Tyrosine phosphorylated:
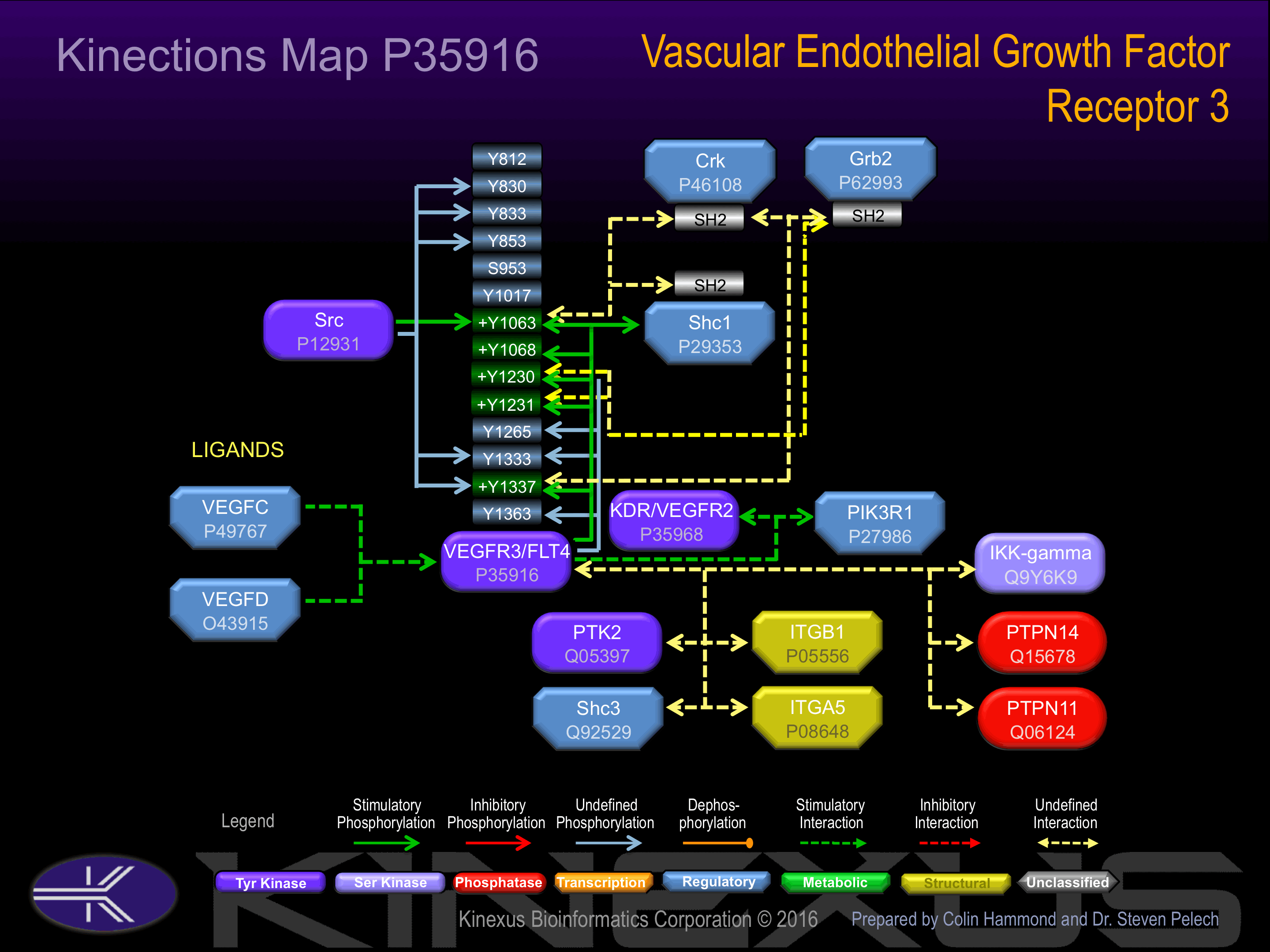
Y830, Y833, Y853, Y1063+, Y1068+, Y1230, Y1231, Y1333, Y1337+, Y1363.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 93
93
913
31
945
 2
2
23
12
16
 13
13
130
29
182
 19
19
191
123
598
 63
63
619
31
449
 0.9
0.9
9
55
13
 14
14
136
39
392
 30
30
292
50
528
 23
23
230
10
227
 11
11
108
122
149
 10
10
97
44
170
 48
48
476
126
484
 11
11
111
40
190
 1
1
13
9
19
 13
13
128
41
246
 5
5
49
18
48
 30
30
290
198
2583
 12
12
118
36
205
 4
4
43
107
53
 43
43
418
112
357
 8
8
83
40
118
 12
12
121
42
185
 9
9
91
38
111
 7
7
64
36
99
 14
14
134
40
241
 65
65
635
91
1156
 7
7
66
43
113
 11
11
113
36
188
 8
8
81
36
115
 9
9
86
42
85
 67
67
660
30
429
 100
100
983
37
1678
 26
26
255
95
995
 66
66
651
83
610
 8
8
80
66
110
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 90.6
90.6
91.7
99 90.6
90.6
91.7
95 -
-
-
86.5 -
-
-
- 91.8
91.8
93
88.5 85.1
85.1
89
- -
-
-
87.5 84.5
84.5
88.7
87 84.3
84.3
88.2
- 42.6
42.6
50.5
- 42.6
42.6
50.5
71 41.4
41.4
58.7
65 26
26
43
57 52.1
52.1
67.4
- 27.1
27.1
43.2
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Activated by binding vascular endothelial growth factor (VEGF), which induces dimerization and autophosphorylation.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
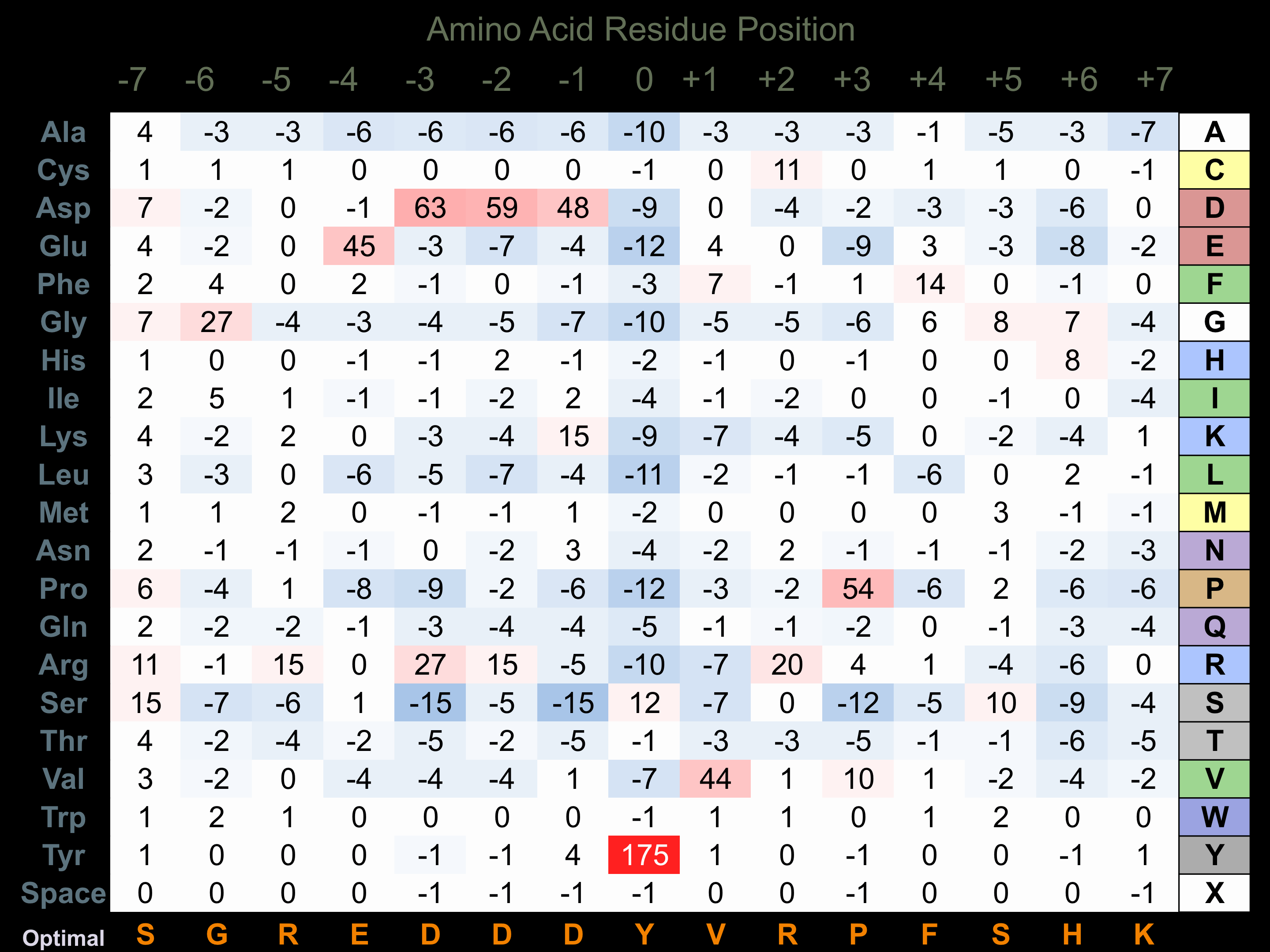
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer, eye, cardiovascular, skin, immune, and bone disorders
Specific Diseases (Non-cancerous):
Hereditary Lymphedema; Lymphedema; Klippel-Trenaunay syndrome; Cellulitis; Intestinal Lymphangiectasia; Hydrocele; Hemifacial Spasm; Intracranial Arteriovenous Malformation
Comments:
Mutations on several sites in FLT4 can attribute to hereditary lymphedema type 1A, which results in that patients with lymphedema suffer from recurrent local infections and physical impairment. P954S and P1137S mutations are associated with hemangioma, capillary infantile. Mutations have been found in family with hereditary lymphedema type IA.
Specific Cancer Types:
Angiosarcomas; Kaposi's sarcomas (KS); Hemangiomas; Lymphangiomas; Hemangioendotheliomas; Hemangiomas, capillary infantile (HCI); Hemangiomas, capillary infantile, somatic; Arteriovenous malformation (AVM); Ovarian carcinosarcomas (OCS); Tufted angioma (TA); Lymphangiosarcomas; Kaposiform hemangioendothelioma (KHE); Lymphatic malformation; Orbital lymphangiomas
Comments:
FLT4 plays critical roles in some lymphangiogenetic mechanisms in cancer and metastasis. It is expressed in acute myeloid leukemia and other types of cancer, and are related to tumour angiogenesis and efficacy of traditional therapies.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain oligodendrogliomas (%CFC= -77, p<0.053); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= -60, p<0.034); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +407, p<0.046); and Prostate cancer - primary (%CFC= +70, p<0.0001). The COSMIC website notes an up-regulated expression score for FLT4 in diverse human cancers of 314, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 1 for this protein kinase in human cancers was 98% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 25708 diverse cancer specimens. This rate is only 39 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.53 % in 1125 large intestine cancers tested; 0.46 % in 806 skin cancers tested; 0.39 % in 590 stomach cancers tested; 0.19 % in 605 oesophagus cancers tested; 0.17 % in 1945 lung cancers tested; 0.08 % in 1396 kidney cancers tested; 0.05 % in 1963 central nervous system cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,903 cancer specimens
Comments:
Ten deletions, 4 insertions and no complex mutations are noted on the COSMIC website.