Nomenclature
Short Name:
NEK3
Full Name:
Serine-threonine-protein kinase Nek3
Alias:
- EC 2.7.11.1
- HSPK 36
- MGC29949
- NimA-related protein kinase 3
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
NEK
SubFamily:
NA
Structure
Mol. Mass (Da):
57,705
# Amino Acids:
506
# mRNA Isoforms:
2
mRNA Isoforms:
57,705 Da (506 AA; P51956); 55,924 Da (489 AA; P51956-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 4 | 257 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S47, S253, S284, S319, S354, S355, S394, S462.
Threonine phosphorylated:
T288, T402, T479.
Tyrosine phosphorylated:
Y273.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 91
91
935
26
1098
 3
3
29
14
29
 6
6
61
2
31
 39
39
396
81
553
 60
60
617
25
571
 100
100
1027
82
3259
 26
26
265
31
430
 68
68
698
29
865
 48
48
498
17
380
 7
7
72
70
53
 5
5
53
19
32
 70
70
719
141
636
 4
4
40
24
14
 3
3
33
12
19
 4
4
44
15
43
 2
2
21
15
13
 19
19
191
110
1803
 3
3
34
9
28
 4
4
41
77
29
 43
43
442
109
469
 6
6
60
13
38
 4
4
37
15
24
 5
5
54
12
13
 9
9
92
11
47
 2
2
16
13
11
 75
75
771
47
1003
 3
3
26
27
14
 4
4
39
11
27
 3
3
32
11
25
 11
11
116
28
57
 60
60
612
36
523
 47
47
478
36
488
 36
36
374
64
708
 69
69
708
57
658
 16
16
165
53
190
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 96.8
96.8
97.2
100 95.4
95.4
96.8
96 -
-
-
87 -
-
-
95 22.4
22.4
31.6
94 -
-
-
- 72.9
72.9
82.9
76 -
-
-
77 -
-
-
- 74.5
74.5
86.3
- 62.2
62.2
76.4
64 21.2
21.2
34.2
58 24.5
24.5
40.7
70 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 26.9
26.9
47.5
38.5 -
-
-
- 23
23
37.5
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CSN2 - P05814 |
| 2 | VAV1 - P15498 |
| 3 | VAV2 - P52735 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
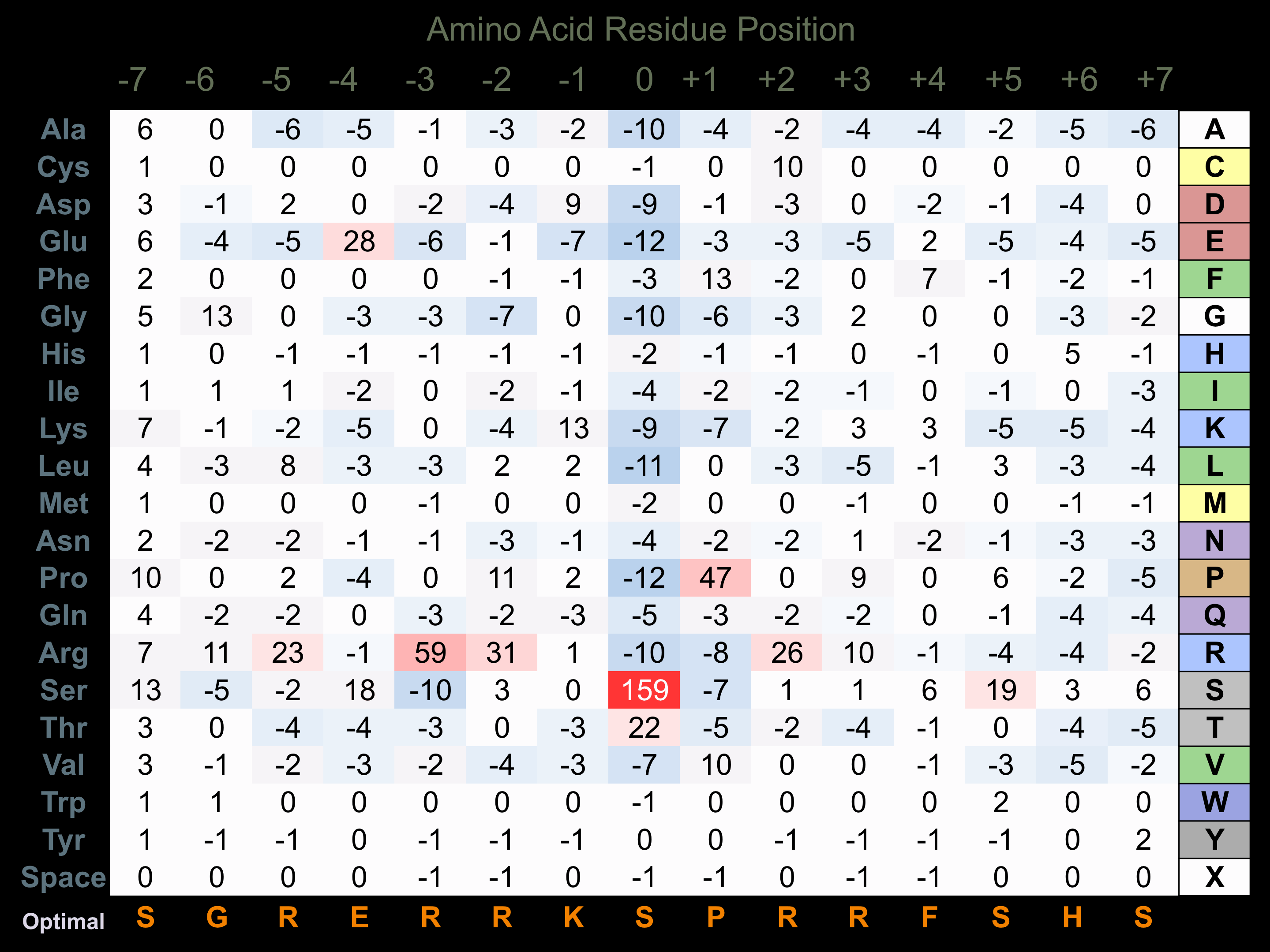
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| AT9283 | IC50 > 100 nM | 24905142 | 19143567 | |
| Cdk1/2 Inhibitor III | IC50 > 150 nM | 5330812 | 261720 | 22037377 |
| MK5108 | IC50 > 500 nM | 24748204 | 20053775 | |
| TG101348 | Kd = 640 nM | 16722836 | 1287853 | 22037378 |
| BI2536 | Kd = 720 nM | 11364421 | 513909 | 22037378 |
| KW2449 | Kd = 1 µM | 11427553 | 1908397 | 22037378 |
| NU6140 | IC50 > 1 µM | 10202471 | 1802728 | 22037377 |
| Silmitasertib | IC50 > 1 µM | 24748573 | 21174434 | |
| SNS314 | IC50 > 1 µM | 16047143 | 514582 | 18678489 |
| Syk Inhibitor | IC50 > 1 µM | 6419747 | 104279 | 22037377 |
| Tpl2 Kinase Inhibitor | IC50 > 1 µM | 9549300 | 22037377 | |
| TWS119 | IC50 > 1 µM | 9549289 | 405759 | 22037377 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Ruxolitinib | Kd = 1.1 µM | 25126798 | 1789941 | 22037378 |
| CHEMBL248757 | Ki > 1.667 µM | 44444843 | 248757 | 17935989 |
| JNJ-28871063 | IC50 < 2 µM | 17747413 | 17975007 | |
| Neratinib | Kd = 2.4 µM | 9915743 | 180022 | 22037378 |
| JNJ-28871063 | IC50 < 3 µM | 17747413 | 17975007 |
Disease Linkage
General Disease Association:
Musculoskeletal disorders
Specific Diseases (Non-cancerous):
Muscle glycogenosis; Phosphorylase kinase deficiency; Muscle phosphorylase kinase deficiency
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +119, p<0.016); Gastric cancer (%CFC= +117, p<0.007); Prostate cancer (%CFC= +148, p<0.068); and Uterine leiomyosarcomas (%CFC= -48, p<0.035).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Frequency of Mutated Sites:
None > 2 in 20,036 cancer specimens

