Nomenclature
Short Name:
PRKY
Full Name:
Serine-threonine-protein kinase PRKY
Alias:
- Protein kinase, Y-linked
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKA
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
31,708
# Amino Acids:
277
# mRNA Isoforms:
1
mRNA Isoforms:
31,708 Da (277 AA; O43930)
4D Structure:
NA
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 49 | 277 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 94
94
869
26
963
 4
4
40
12
26
 9
9
88
2
65
 30
30
283
74
402
 70
70
654
22
570
 6
6
58
74
61
 23
23
213
28
421
 59
59
551
24
481
 40
40
371
14
198
 8
8
70
70
51
 6
6
60
18
36
 77
77
711
139
586
 11
11
100
24
67
 5
5
48
9
12
 8
8
74
12
38
 3
3
30
15
19
 9
9
85
106
43
 6
6
60
8
34
 5
5
46
74
24
 56
56
516
106
521
 15
15
140
10
78
 15
15
137
14
137
 6
6
60
4
28
 10
10
89
8
33
 16
16
149
10
151
 84
84
779
40
713
 8
8
77
24
53
 10
10
92
8
37
 7
7
66
8
43
 17
17
154
28
69
 39
39
366
12
195
 69
69
645
30
536
 4
4
37
34
34
 100
100
929
52
794
 12
12
109
26
56
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 72.3
72.3
74.9
0 71
71
73.7
- -
-
-
- -
-
-
- 61.6
61.6
66.6
- -
-
-
- 56.3
56.3
64.2
- 37.9
37.9
54.4
- -
-
-
- 49.8
49.8
55.1
- 31.3
31.3
43.1
- -
-
-
- 30
30
44.1
- -
-
-
- 28.1
28.1
38.6
- 42.5
42.5
54
- 32.9
32.9
51
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 36
36
50.5
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
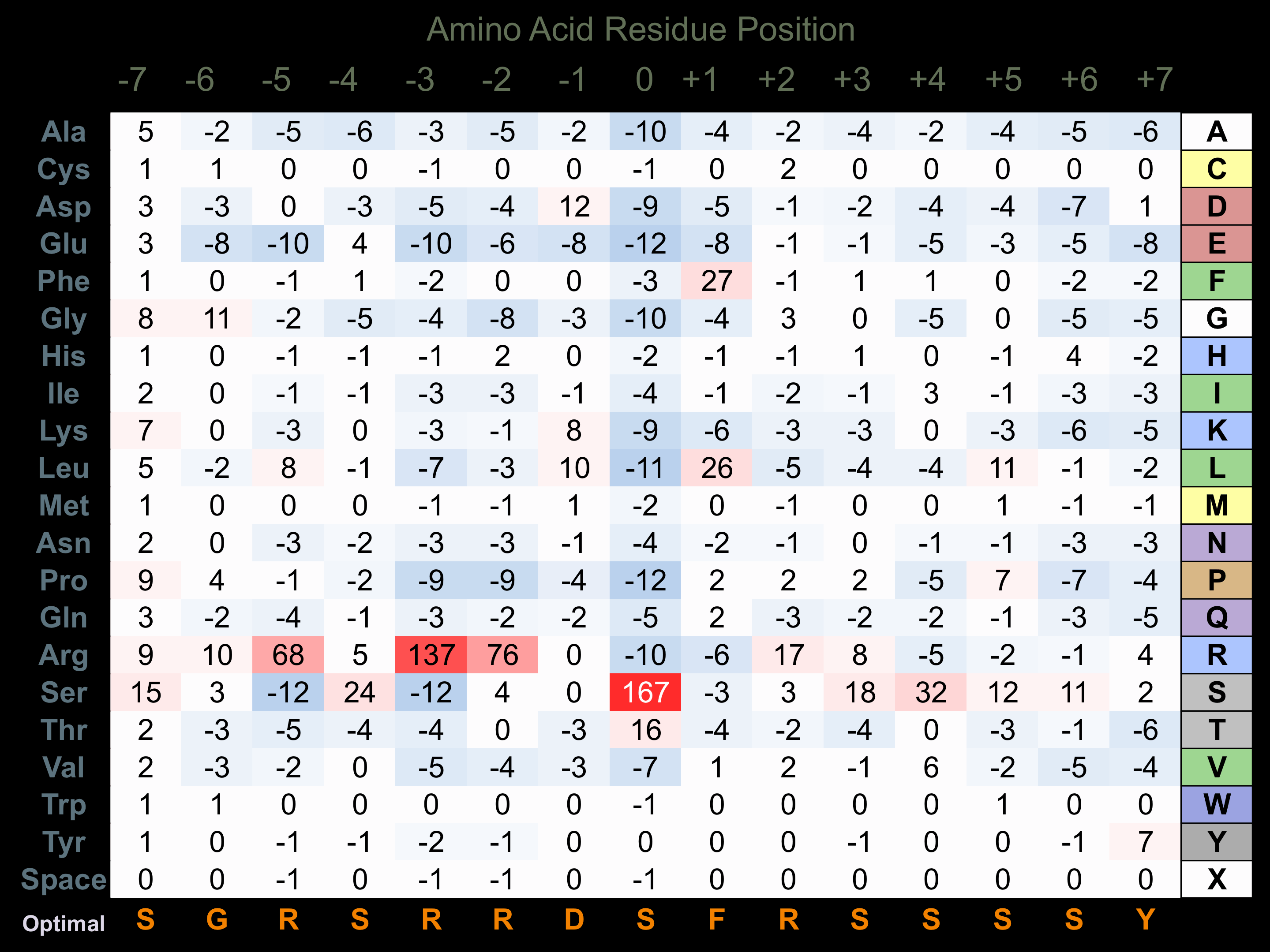
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Development disorders
Specific Diseases (Non-cancerous):
Sex reversal disorder
Comments:
A chromosomal aberration associating with PRKY is a cause of sex reversal disorder leading to XX males and XY females, which accounts for about 30% of the cases of sex reversal disorder.
Comments:
PRKY may be a tumour requiring protein (TRP), since it displays extremely low rates of mutation in human cancers.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Ovary adenocarcinomas (%CFC= +88, p<0.028).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.02 % in 24825 diverse cancer specimens. This rate is -79 % lower than the average rate of 0.075 % calculated for human protein kinases in general. Such a very low frequency of mutation in human cancers is consistent with this protein kinase playing a role as a tumour requiring protein (TRP).
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.17 % in 864 skin cancers tested; 0.06 % in 589 stomach cancers tested; 0.06 % in 558 thyroid cancers tested; 0.06 % in 1823 lung cancers tested.
Frequency of Mutated Sites:
None > 2 in 20,108 cancer specimens
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website. In human tumours, almost of the point mutations are silent or conserved amino acid substitutions with no insertions, deletions or complex mutations.

