Nomenclature
Short Name:
PKG2
Full Name:
cGMP-dependent protein kinase 2
Alias:
- cGK 2
- PRKG2
- PRKGR2
- Protein kinase, cGMP-dependent, type II
- Type II cGMP-dependent protein kinase
- cGKII
- EC 2.7.11.12
- KGP2
- Kinase cGMP-dependent protein kinase 2PKG2
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
PKG
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
87,432
# Amino Acids:
762
# mRNA Isoforms:
2
mRNA Isoforms:
87,432 Da (762 AA; Q13237); 84,398 Da (733 AA; Q13237-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 19 | 85 | Coiled-coil |
| 168 | 284 | cNMP |
| 286 | 409 | cNMP |
| 453 | 711 | Pkinase |
| 712 | 762 | Pkinase_C |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Methylated:
K521.
Myristoylated:
G2.
Serine phosphorylated:
S11, S92, S110, S114, S117, S126, S445, S448.
Threonine phosphorylated:
T109, T131, T609+.
Tyrosine phosphorylated:
Y306, Y307, .
Ubiquitinated:
K157, K165.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 85
85
1016
16
1400
 4
4
52
10
76
 3
3
39
14
62
 35
35
419
62
1650
 29
29
345
13
314
 4
4
54
42
120
 17
17
209
19
286
 44
44
531
38
985
 36
36
432
10
427
 4
4
51
57
66
 3
3
37
29
55
 41
41
489
112
614
 2
2
27
29
34
 4
4
50
7
57
 3
3
34
11
44
 2
2
28
8
22
 2
2
29
116
44
 3
3
33
25
57
 3
3
40
60
41
 23
23
274
56
304
 3
3
38
27
62
 1.4
1.4
17
29
27
 2
2
28
26
41
 3
3
33
24
63
 2
2
26
27
42
 100
100
1202
43
2778
 2
2
23
26
28
 3
3
33
25
51
 2
2
27
24
44
 11
11
135
14
46
 33
33
397
18
271
 87
87
1040
21
1941
 58
58
694
47
1057
 51
51
614
31
637
 2
2
24
22
23
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 92.6
92.6
93.2
100 87.7
87.7
87.9
100 -
-
-
97 -
-
-
- 96.7
96.7
98.6
97 -
-
-
- 96
96
98.6
97 96.3
96.3
99.3
96 -
-
-
- -
-
-
- -
-
-
81 70.7
70.7
82.7
72 69.5
69.5
83.8
- -
-
-
- 45.4
45.4
64.9
52 -
-
-
- 45
45
64.2
49 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 22.6
22.6
38
- 22
22
35.7
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PTS - Q03393 |
| 2 | PLCB3 - Q01970 |
| 3 | MYLK - Q15746 |
Regulation
Activation:
Binding of cGMP stimulates phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| eNOS | P29474 | S1177 | TSRIRTQSFSLQERQ | + |
| eNOS | P29474 | S633 | WRRKRKESSNTDSAG | |
| eNOS | P29474 | T495 | TGITRKKTFKEVANA | |
| GCAP2 | Q9UMX6 | S197 | LAQQRRKSAMF____ | |
| ICLN | P54105 | S2 | ______MSFLKSFPP | |
| ICLN | P54105 | S45 | YIAESRLSWLDGSGL | |
| Lasp-1 | Q14847 | S146 | MEPERRDSQDGSSYR | |
| PKG2 (PRKG2) | Q13237 | S110 | LEVHRKTSGLVSLHS | |
| PKG2 (PRKG2) | Q13237 | S114 | RKTSGLVSLHSRRGA | |
| PKG2 (PRKG2) | Q13237 | S117 | SGLVSLHSRRGAKAG | |
| PKG2 (PRKG2) | Q13237 | S126 | RGAKAGVSAEPTTRT | |
| PKG2 (PRKG2) | Q13237 | S445 | KEKVARFSSSSPFQN | |
| PKG2 (PRKG2) | Q13237 | S92 | VVHMQGGSPLQASPD | |
| PKG2 (PRKG2) | Q13237 | T109 | PLEVHRKTSGLVSLH | |
| PLCB3 | Q01970 | S1105 | LDRKRHNSISEAKMR | - |
| PLCB3 | Q01970 | S26 | VETLRRGSKFIKWDE | - |
| PTS | Q03393 | S19 | AQVSRRISFSASHRL | |
| StAR | P49675 | S100 | QEGWKKESQQDNGDK | |
| StAR | P49675 | S56 | INQVRRRSSLLGSRL | |
| StAR | P49675 | S57 | NQVRRRSSLLGSRLE |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
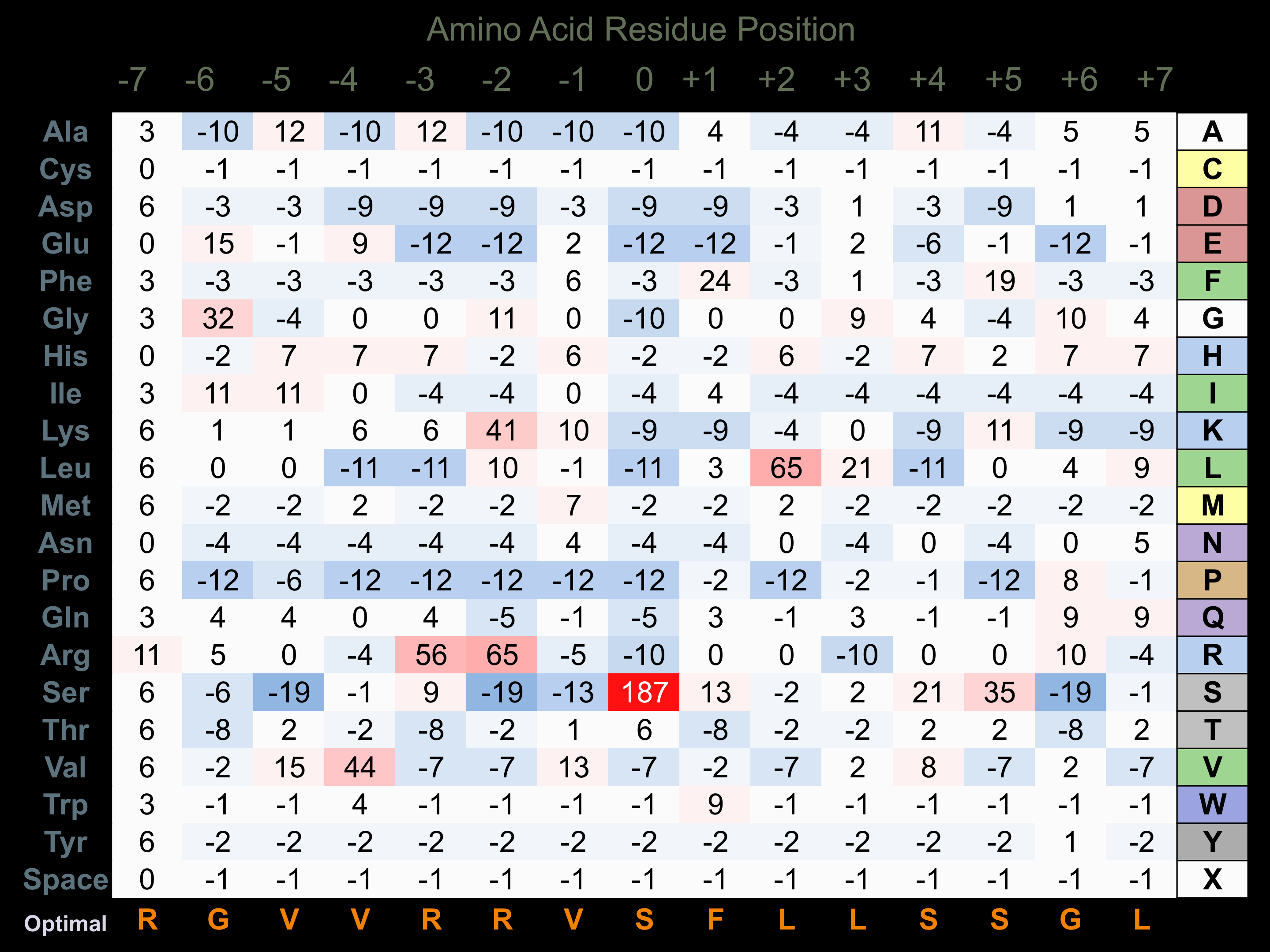
Matrix Type:
Experimentally derived from alignment of 30 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, amino acid metabolism disorders
Specific Diseases (Non-cancerous):
Hyperphenylalaninemia (HPA)
Comments:
Hyperphenylalaninemia (HPA) is a rare disorder arising from an excessive amount of phenylalanine circulating in the blood. HPA can lead to phenylketonuria which can cause cognitive impairment and seizures.
Specific Cancer Types:
Colon cancer
Comments:
PKG2 could have a role in treating colon cancers.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -55, p<0.004); Colon mucosal cell adenomas (%CFC= -67, p<0.0001); Colorectal adenocarcinomas (early onset) (%CFC= -50, p<0.031); and Ovary adenocarcinomas (%CFC= +131, p<0.008).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 24433 diverse cancer specimens. This rate is only 37 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.58 % in 864 skin cancers tested; 0.44 % in 1229 large intestine cancers tested; 0.35 % in 555 stomach cancers tested; 0.29 % in 273 cervix cancers tested; 0.26 % in 603 endometrium cancers tested; 0.17 % in 548 urinary tract cancers tested; 0.17 % in 1608 lung cancers tested; 0.11 % in 238 bone cancers tested; 0.09 % in 710 oesophagus cancers tested; 0.09 % in 1512 liver cancers tested; 0.09 % in 1316 breast cancers tested; 0.07 % in 2103 central nervous system cancers tested; 0.07 % in 1253 kidney cancers tested; 0.05 % in 807 ovary cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S143F (5).
Comments:
Only 3 deletions, 2 insertions, and no complex mutations are noted on the COSMIC website.

