Nomenclature
Short Name:
ADCK1
Full Name:
AarF domain containing kinase 1
Alias:
- LOC57143,EC 2.7.11.-
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
ABC1
SubFamily:
ABC1-B
Structure
Mol. Mass (Da):
51,999
# Amino Acids:
455
# mRNA Isoforms:
3
mRNA Isoforms:
60,577 Da (530 AA; Q86TW2); 59,719 Da (523 AA; Q86TW2-2); 51,999 Da (455 AA; Q86TW2-3)
4D Structure:
NA
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 17 | signal_peptide |
| 80 | 455 | ABC1 |
| Pkinase |
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 70
70
1227
9
919
 1.5
1.5
26
4
32
 2
2
37
8
43
 7
7
123
40
84
 42
42
741
13
620
 4
4
71
9
44
 2
2
41
13
25
 11
11
187
16
515
 0.5
0.5
8
3
2
 8
8
136
37
102
 2
2
27
12
42
 28
28
489
19
676
 2
2
39
8
39
 0.3
0.3
5
3
1
 4
4
72
12
80
 3
3
58
5
13
 7
7
121
76
75
 2
2
39
11
46
 7
7
122
29
93
 45
45
780
31
484
 4
4
63
13
69
 3
3
48
13
61
 2
2
37
7
39
 1.3
1.3
23
11
29
 5
5
88
13
109
 32
32
563
31
720
 2
2
32
11
39
 2
2
31
11
34
 2
2
27
10
35
 8
8
141
14
70
 69
69
1207
12
70
 100
100
1752
11
5399
 21
21
375
42
620
 58
58
1021
26
842
 7
7
123
22
72
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 79.3
79.3
79.5
100 97.1
97.1
98.1
98.5 -
-
-
88 -
-
-
- 58.1
58.1
63
92.5 -
-
-
- 87.3
87.3
93
88.5 74.1
74.1
80.1
86 -
-
-
- 54.7
54.7
63.4
- 75.2
75.2
87.7
78 62.6
62.6
79.4
64 69
69
81.1
71 -
-
-
- 44.7
44.7
65.4
49 45.8
45.8
64.3
- -
-
-
45 -
-
-
- -
-
-
- -
-
-
- -
-
-
44 36.8
36.8
58.7
42 30
30
49
36 -
-
-
38
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
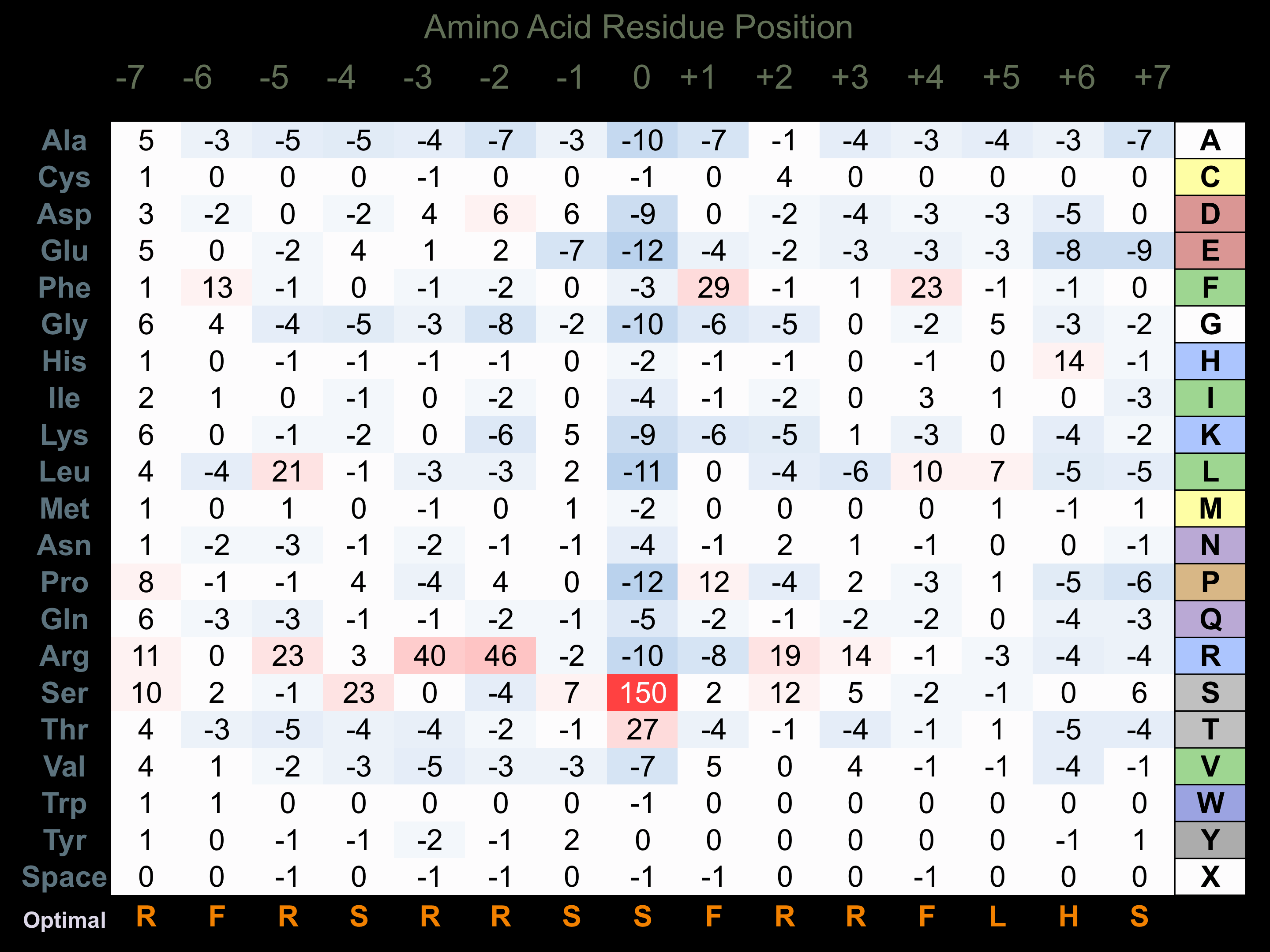
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Ovary adenocarcinomas (%CFC= +71, p<0.029); Pituitary adenomas (aldosterone-secreting) (%CFC= +51, p<0.008); and Vulvar intraepithelial neoplasias (%CFC= -57, p<0.025). The COSMIC website notes an up-regulated expression score for ADCK1 in diverse human cancers of 416, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 58 for this protein kinase in human cancers was similar to the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 24726 diverse cancer specimens. This rate is only 21 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.51 % in 864 skin cancers tested; 0.38 % in 1270 large intestine cancers tested; 0.3 % in 589 stomach cancers tested; 0.29 % in 603 endometrium cancers tested.
Frequency of Mutated Sites:
None >2 in 19643 cancer specimens
Comments:
Only 1 deletion mutation at the C-terminal end of the protein are listed on the COSMIC website.

