Nomenclature
Short Name:
KSR
Full Name:
Kinase suppressor of ras-1
Alias:
- CAP kinase
- Ceramide-activated protein kinase
- Kinase suppressor of RAS-1
- RSU2
- HB
- HB protein
- KSR1
- Kinase suppressor of ras 1
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
RAF
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
102,032
# Amino Acids:
921
# mRNA Isoforms:
4
mRNA Isoforms:
102,160 Da (923 AA; Q8IVT5); 97,115 Da (877 AA; Q8IVT5-2); 87,344 Da (786 AA; Q8IVT5-3); 84,697 Da (762 AA; Q8IVT5-4)
4D Structure:
Interacts with HSPCA/HSP90, YWHAB/14-3-3, CDC37, MAP2K/MEK, MARK3, PPP2R1A and PPP2CA. Also interacts with RAF and MAPK/ERK, in a Ras-dependent manner By similarity. The binding of 14-3-3 proteins to phosphorylated KSR prevents the membrane localization.
1D Structure:
Subfamily Alignment

Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Threonine phosphorylated:
T238, T240, T248, T250, T261, T268, T270, T271, T272, T273, T274, T286, T288, T402, T404, T423, T425, T471, T473, T597, T599, T844.
Tyrosine phosphorylated:
Y160, Y162, Y721, Y723.
Acetylated:
K656, K709.
Serine phosphorylated:
S182, S184, S202, S227, S229, S236, S238, S244, S255, S257, S265, S267, S291, S293, S307, S309, S311, S312, S314, S329, S331, S332, S334, S351, S404, S406, S407, S409, S443, S447, S456, S476, S478, S567, S569, S595, S597, S886, S888.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 61
61
972
51
1333
 2
2
39
19
44
 0.6
0.6
9
3
2
 21
21
336
147
571
 38
38
605
41
478
 2
2
28
123
116
 14
14
221
54
477
 100
100
1583
47
2628
 20
20
324
21
215
 11
11
167
137
313
 3
3
43
31
56
 40
40
628
239
617
 1
1
16
36
14
 2
2
25
12
23
 14
14
216
20
428
 3
3
46
27
72
 2
2
38
209
159
 3
3
47
11
28
 7
7
104
132
204
 26
26
415
187
494
 8
8
125
19
191
 6
6
96
25
184
 2
2
36
14
16
 3
3
55
11
47
 7
7
107
19
181
 99
99
1560
85
2829
 0.9
0.9
15
36
19
 3
3
43
11
42
 1.3
1.3
20
11
15
 6
6
101
56
85
 19
19
293
30
353
 39
39
611
56
664
 6
6
93
96
248
 54
54
861
109
796
 6
6
92
52
118
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 40.4
40.4
55.8
97 98.3
98.3
98.7
99 -
-
-
94 -
-
-
40 89.1
89.1
91.2
93.5 -
-
-
- 86.4
86.4
89.4
91 22.8
22.8
36.8
94 -
-
-
- 20.4
20.4
24.8
- 24.1
24.1
38.5
77 23.1
23.1
37.9
77 43.3
43.3
55.7
66 -
-
-
- 24.5
24.5
41.5
33 31.7
31.7
47.2
- 20.9
20.9
36.9
30 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MAP2K1 - Q02750 |
| 2 | RAF1 - P04049 |
| 3 | MAP2K2 - P36507 |
| 4 | NME1 - P15531 |
| 5 | NME2 - P22392 |
| 6 | BRAP - Q7Z569 |
| 7 | YWHAH - Q04917 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ERK2 | P28482 | T272 | SFITPPTTPQLRRHT | |
| ERK2 | P28482 | T286 | TKLKPPRTPPPPSRK | |
| MARK3 | P27448 | S309 | PTLTRSKSHESQLGN | |
| MARK3 | P27448 | S404 | TRLRRTESVPSDINN | |
| TAK1 | O43318 | S404 | TRLRRTESVPSDINN | |
| NM23 | P15531 | S404 | TRLRRTESVPSDINN | |
| ERK2 | P28482 | S443 | AMNHLDSSSNPSSTT | |
| ERK1 | P27361 | S443 | AMNHLDSSSNPSSTT | |
| NM23 | P15531 | S447 | LDSSSNPSSTTSSTP | |
| ERK2 | P28482 | S456 | TTSSTPSSPAPFPTS |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
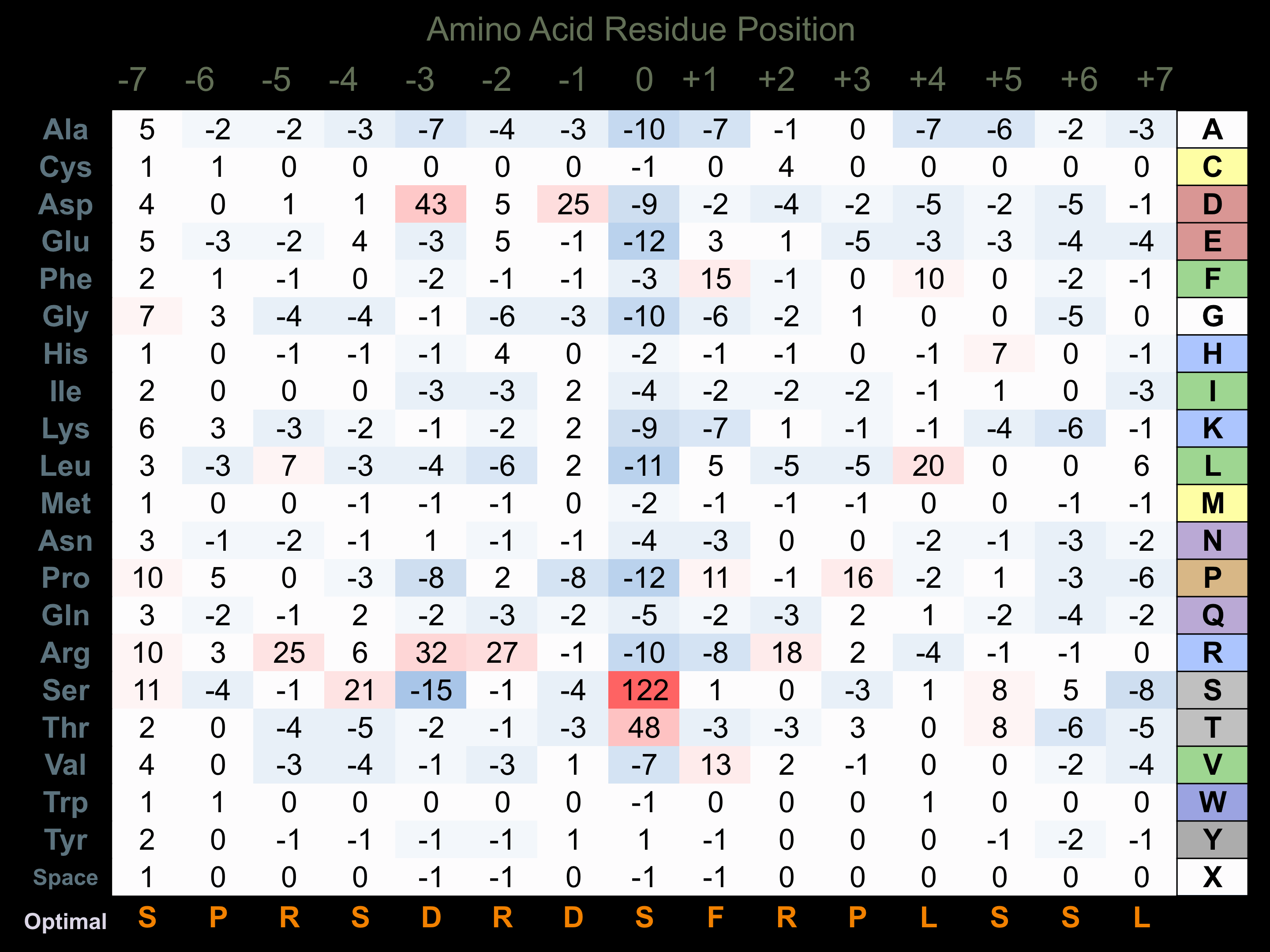
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Respiratory disorders, and infectious disease
Specific Diseases (Non-cancerous):
Pharyngitis; Bronchopneumonia; Scarlet fever
Comments:
Not much correlations of the gene with human diseases have been found. However, in animal studies, KSR was found to increase suspectibility to chronic colitis and apoptosis of mouse colon epithelial cells. KSR knock-out mice were highly susceptible to pulmonary Pseudomonas aeruginosa infection and may cause death of mice, while normal mice cleared the infection.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -45, p<0.009); Cervical cancer stage 1B (%CFC= -82); Cervical cancer stage 2A (%CFC= -61); Classical Hodgkin lymphomas (%CFC= +51, p<0.022); Clear cell renal cell carcinomas (cRCC) (%CFC= +177, p<0.019); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +5857, p<0.005).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24136 diverse cancer specimens. This rate is the same as the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
None > 3 in 20,827 cancer specimens
Comments:
Nineteen deletions (all P291fs*36) and 5 insertions (three P292fs*22 and two P292fs*36), and no complex mutations noted on the COSMIC website.

