Nomenclature
Short Name:
VRK1
Full Name:
Serine-threonine-protein kinase VRK1
Alias:
- EC 2.7.11.1
- Serine/threonine protein kinase VRK1
- Vaccinia related kinase 1
- Vaccinia-related kinase 1
Classification
Type:
Protein-serine/threonine kinase
Group:
CK1
Family:
VRK
SubFamily:
NA
Structure
Mol. Mass (Da):
45,476
# Amino Acids:
396
# mRNA Isoforms:
1
mRNA Isoforms:
45,476 Da (396 AA; Q99986)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 37 | 325 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K140, K147.
Serine phosphorylated:
S75, S158, S342, S376+, S432+.
Threonine phosphorylated:
T153, T355, T378, T380.
Ubiquitinated:
K16, K112, K188, K301, K329.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 34
34
692
19
1094
 0.8
0.8
17
9
26
 16
16
325
13
259
 29
29
585
58
1343
 38
38
774
14
755
 55
55
1108
45
4264
 15
15
301
19
459
 17
17
355
29
405
 25
25
515
10
514
 3
3
70
57
113
 8
8
170
26
257
 23
23
466
117
554
 16
16
320
24
364
 3
3
60
9
60
 6
6
120
23
156
 2
2
36
8
22
 23
23
458
114
3765
 14
14
288
19
354
 3
3
60
58
113
 31
31
626
56
679
 9
9
187
22
250
 14
14
284
23
330
 15
15
295
22
401
 36
36
725
20
642
 33
33
662
22
689
 30
30
618
37
603
 13
13
270
27
309
 11
11
221
20
322
 12
12
243
20
274
 5
5
103
14
63
 51
51
1027
18
657
 100
100
2033
21
2571
 71
71
1439
55
1486
 39
39
798
31
747
 3
3
67
22
69
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
99.8
100 98.2
98.2
99
98 -
-
-
91 -
-
-
93 83.8
83.8
88.1
90 -
-
-
- 76.8
76.8
83.4
86 29
29
46.5
88 -
-
-
- 66.2
66.2
73.1
- 70.2
70.2
81.6
73 47.3
47.3
59.4
69 59.3
59.3
72.5
64 -
-
-
- 31.1
31.1
43.7
37 -
-
-
- -
-
-
35.5 39.8
39.8
56
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 20.9
20.9
37.4
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | TP53 - P04637 |
| 2 | ATF2 - P15336 |
| 3 | PLK3 - Q9H4B4 |
| 4 | GOLGB1 - Q14789 |
| 5 | VRK3 - Q8IV63 |
| 6 | RCC1 - P18754 |
| 7 | RAN - P62826 |
Regulation
Activation:
Active in presence of Mn2+, Mg2+ and Zn2+.
Inhibition:
not functional with Ca2+ or Cu2+.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ATF2 | P15336 | S62 | FGPARNDSVIVADQT | + |
| ATF2 | P15336 | T116 | LDLSPLATPIIRSKI | + |
| ATF2 | P15336 | T73 | ADQTPTPTRFLKNCE | + |
| CREB1 | P16220 | S133 | EILSRRPSYRKILND | + |
| H3.1 | P68431 | S11 | TKQTARKSTGGKAPR | + |
| H3.1 | P68431 | T4 | ____MARTKQTARKS | + |
| Jun (c-Jun) | P05412 | S63 | KNSDLLTSPDVGLLK | + |
| Jun (c-Jun) | P05412 | S73 | VGLLKLASPELERLI | + |
| p53 | P04637 | T18 | EPPLSQETFSDLWKL | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
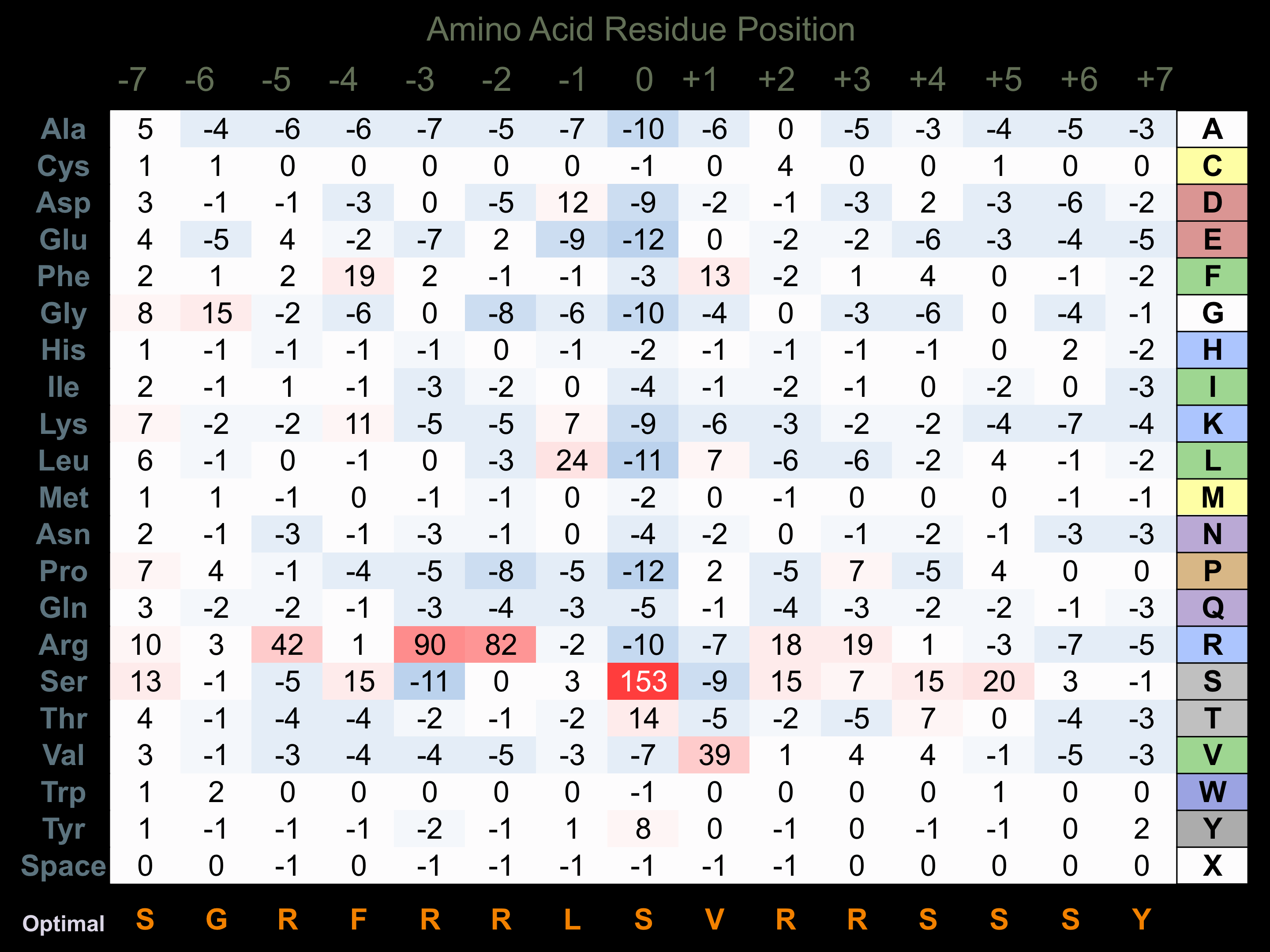
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Ro-31-8220 | IC50 = 34 nM | 5083 | 6291 | 21829721 |
| Staurosporine | IC50 > 37 nM | 5279 | 21829721 | |
| IC261 | IC50 > 56 nM | 3674 | 576349 | 21829721 |
| PKR Inhibitor | IC50 > 1 µM | 6490494 | 235641 | 22037377 |
| SureCN4875304 | IC50 > 3.5 µM | 46871765 | 20472445 |
Disease Linkage
General Disease Association:
Musculoskeletal, and infectious disorder
Specific Diseases (Non-cancerous):
Vaccinia; Pontocerebellar hypoplasia (PCH1A); Werdnig-Hoffmann disease; Anterior horn cell disease
Comments:
Mutations of VRK are associated with pontocerebellar hypoplasia 1A, which is a disorder characterized by an abnormally small cerebellum and brainstem, central and peripheral motor dysfunction from birth, and severe intellectual disability.
Comments:
VRK1 was showed to confer resistance to DNA-damaging in human breast cancer.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +113, p<0.008); Bladder carcinomas (%CFC= +192, p<0.0002); Cervical cancer (%CFC= -63, p<0.0001); Colon mucosal cell adenomas (%CFC= +127, p<0.0001); Large B-cell lymphomas (%CFC= +51, p<0.008); Lung adenocarcinomas (%CFC= +47, p<0.004); Oral squamous cell carcinomas (OSCC) (%CFC= +66, p<0.005); Ovary adenocarcinomas (%CFC= +91, p<0.039); Prostate cancer (%CFC= +100, p<0.061); and Vulvar intraepithelial neoplasia (%CFC= +132, p<0.0001). The COSMIC website notes an up-regulated expression score for VRK1 in diverse human cancers of 564, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 31 for this protein kinase in human cancers was 0.5-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24751 diverse cancer specimens. This rate is only -28 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.42 % in 602 endometrium cancers tested; 0.35 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
None > 2 in 20,034 cancer specimens
Comments:
Only 3 deletions, and no insertions or complex mutations are noted on the COSMIC website.

