Nomenclature
Short Name:
NRBP1
Full Name:
Multiple domain putative nuclear protein
Alias:
- BCON3
- DKFZp564D1878
- Nuclear receptor binding protein
- FLJ11084
- MADM
- MUDPNP
- NRBP
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
NRBP
SubFamily:
NA
Structure
Mol. Mass (Da):
59,845
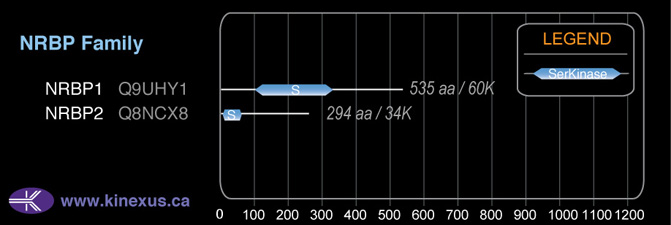
# Amino Acids:
535
# mRNA Isoforms:
1
mRNA Isoforms:
59,845 Da (535 AA; Q9UHY1)
4D Structure:
Homodimer. Binds to MLF1, recruiting a serine kinase which phosphorylates both itself and MLF1. Phosphorylated MLF1 binds to YWHAZ and is retained in the cytoplasm By similarity.
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 68 | 327 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K134.
Serine phosphorylated:
S2, S6, S11, S12, S14, S49, S363, S422.
Threonine phosphorylated:
T8, T225, T232, T421, T431, T433.
Tyrosine phosphorylated:
Y152.
Ubiquitinated:
K517.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 19
19
844
16
1043
 9
9
395
10
730
 19
19
850
18
690
 15
15
697
75
1031
 20
20
882
13
778
 5
5
208
45
320
 2
2
78
15
29
 34
34
1537
48
2137
 15
15
669
10
600
 9
9
404
52
595
 13
13
573
31
608
 20
20
909
93
654
 13
13
570
31
608
 10
10
467
8
926
 13
13
570
11
816
 8
8
368
9
561
 31
31
1395
102
4631
 12
12
545
25
457
 10
10
443
56
616
 15
15
688
58
677
 31
31
1395
27
3392
 12
12
549
27
490
 18
18
793
19
646
 26
26
1187
27
1173
 13
13
578
27
435
 24
24
1070
56
1145
 12
12
540
34
614
 13
13
596
25
468
 26
26
1180
25
1092
 3
3
142
14
40
 21
21
959
18
616
 100
100
4519
23
8199
 5
5
236
55
319
 20
20
904
26
680
 9
9
405
22
494
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 79.7
79.7
79.7
100 98.5
98.5
98.5
100 -
-
-
100 -
-
-
100 73.8
73.8
73.8
100 -
-
-
- 98.3
98.3
99.2
98 -
-
-
99 -
-
-
- 91.4
91.4
94.5
- 87.6
87.6
91.7
88 86.1
86.1
91.9
88 82.6
82.6
89.5
84 -
-
-
- 45.2
45.2
60.7
53 55
55
66.7
- -
-
-
57 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 24.4
24.4
42.9
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MLF1 - P58340 |
| 2 | RAC3 - P60763 |
| 3 | TSC22D4 - Q9Y3Q8 |
| 4 | ADAM10 - O14672 |
| 5 | TSC22D1 - Q15714 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
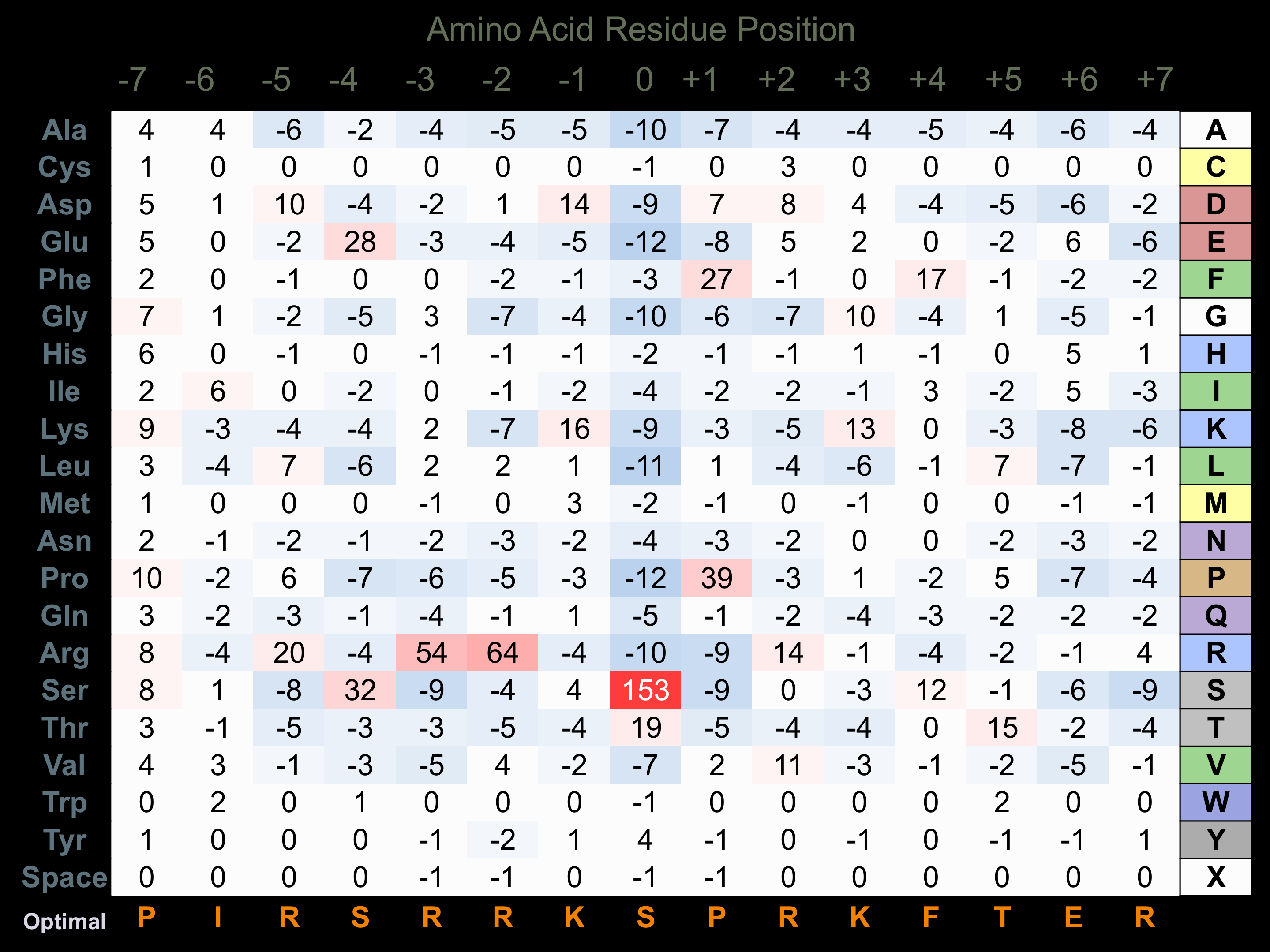
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Prostate cancer
Comments:
High expression of NRBP1 in prostate cancer is associated with poor clinical outcomes and enhanced cancer growth. It also was shown to regulate homeostasis and tumour formation of intestinal progenitor cells.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -56, p<0.001); Bladder carcinomas (%CFC= +65, p<0.0004); Breast epithelial cell carcinomas (%CFC= +89, p<0.001); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= -45, p<0.024); Cervical cancer (%CFC= +55, p<0.002); Skin fibrosarcomas (%CFC= +123); Skin melanomas - malignant (%CFC= +126, p<0.0004); and Uterine fibroids (%CFC= +49, p<0.016). The COSMIC website notes an up-regulated expression score for NRBP1 in diverse human cancers of 616, which is 1.3-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 135 for this protein kinase in human cancers was 2.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24726 diverse cancer specimens. This rate is only -18 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.38 % in 1270 large intestine cancers tested; 0.19 % in 603 endometrium cancers tested; 0.19 % in 589 stomach cancers tested; 0.14 % in 273 cervix cancers tested; 0.1 % in 548 urinary tract cancers tested; 0.09 % in 864 skin cancers tested; 0.09 % in 1512 liver cancers tested; 0.09 % in 1316 breast cancers tested; 0.07 % in 1634 lung cancers tested; 0.07 % in 1276 kidney cancers tested; 0.04 % in 881 prostate cancers tested; 0.04 % in 833 ovary cancers tested; 0.03 % in 710 oesophagus cancers tested; 0.03 % in 1459 pancreas cancers tested; 0.02 % in 942 upper aerodigestive tract cancers tested; 0.01 % in 2103 central nervous system cancers tested; 0.01 % in 2009 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R506Q (3).
Comments:
Only 5 deletions, no insertions or complex mutations are noted on the COSMIC website.

